 |
| welcome/ |
| java-mcmc/ |
| software/ |
| papers/ |
| links/ |
| email me |

|
The applet on this page demonstrates how to produce perfect samples for a general class of Bayesian classification
problems. Consider a dataset where the points 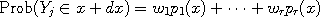
where the probability densities According to Bayesian principles, there exists a unique well defined posterior probability distribution which represents our degree of belief about the values of the various weights, and methods such as the Data Augmentation algorithm can be used to approximately produce random samples from it.
The basic idea for this consists in randomly allocating each data point With Perfect Simulation methodology, we can draw the weights directly from the posterior probabilities, in a finite time. The following paper explains how to do this, and also generalizes the above problem to the case of unknown means. Below, I only give enough information to understand the applet.
Once you press the button above to start the applet, you will see a new window which is split into two halves. The top panel
on the left side shows the available data points
In the middle panel, you see an evolving blue line. At each sweep, this gets updated to reflect a randomly adjusted allocation
of datapoints to each component. Since there are three components to begin with, the blue line is formed of three segments.
The height of each segment is proportional to the number of data points among
Think of an allocation as an opinion about the dataset. The Gibbs sampler perturbs this opinion slightly, in a manner
consistent with the posterior distribution on the unknown weights In the applet, every possible opinion could be drawn besides the blue line we see, but this would be very confusing. Instead, we sandwich the other opinions between a yellow and an orange region. Once these two meet, we know that all possible initial opinions about the allocations have finally agreed. We budget 100 sweeps by default for each coalescence. At the end of these, we either have a coalescence or we don't, but we start again with all possible opinions.
The bottom panel shows some statistics about the perfect simulation method, and also draws a color coded histogram of the obtained weights. With enough IID samples, this gives a representation of the posterior distribution for each weight. |



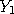 ,...,
,...,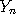 are assumed to follow a probability distribution of the form
are assumed to follow a probability distribution of the form
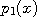 ,...,
,...,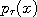 are already known,
but the weighting factors
are already known,
but the weighting factors 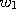 ,...,
,...,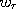 are not. To fix ideas, think of the
are not. To fix ideas, think of the
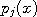 as Gaussians with known means and variances.
The problem is to infer the weights from the available data.
as Gaussians with known means and variances.
The problem is to infer the weights from the available data.
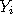 to some component while preserving the posterior probabilities. A Gibbs sampling algorithm is used to dynamically update the allocations (and hence the weights) in a random but self consistent manner. In the long run, this produces weights which approximately follow the posterior probabilities.
to some component while preserving the posterior probabilities. A Gibbs sampling algorithm is used to dynamically update the allocations (and hence the weights) in a random but self consistent manner. In the long run, this produces weights which approximately follow the posterior probabilities.